Principal Component Analysis (PCA) is one of the most famous models used for dimensionality reduction. It uses an orthonormal transformation to convert a set of observation of possibly correlated variables into a set of linearly uncorrelated variables. It corresponds to a feature extraction technique, which means that instead of selecting a subset of the original features, these methods transform the original features into a new set of features with reduced dimensionality, in Dimensionality Reduction and Feature Extraction you can read more about dimensionality reduction and feature extraction techniques.
Given a dataset consisting of a set of points (vectors) in a high dimensional space, the main idea of PCA is to find the directions, also called principal components, along which the points line up best and make a projection on these components to create a new reduced dataset, but capturing the most relevant information.
The PCA algorithm starts by choosing the number of principal components (number of dimensions to reduce), then the covariance matrix of the dataset is calculated, as well as the eigenvectors and eigenvalues of this matrix. The eigenvectors are used to perform the transformation and eigenvalues to choose which eigenvectors are taken. The N-eigenvectors with the highest corresponding eigenvalues (in descend order) are used as to build a transformation matrix. Finally, to obtain a low dimensional dataset with new features, the original dataset is multiplied by the transformation matrix. The whole process is described in the next image, which provides an outline of the basic PCA algorithm.
Implementing algorithm in Python
Depending on each dataset, the selection of the number of dimensions may vary. In this post we generate a 2-dimensional dataset and reduce it to a 1-dimensional dataset, but these parameters can be easily modified to apply the PCA algorithm with different datasets.
Importing libraries
First, we import the necessary libraries for the code, which includes:
# Importing libraries
import numpy as np
from numpy.linalg import eig
from sklearn.preprocessing import normalize
from sklearn.decomposition import PCA as PCA_sklearn
from sklearn.preprocessing import StandardScaler
from scipy.stats import multivariate_normal
import matplotlib.pyplot as plt
- numpy: Provides mathematical functions for arrays, we also import the function eig to calculate eigenvalues and eigenvectors.
- sklearn: Library that contains methods for data preprocessing and machine learning models. In this post the functions imported are: StandardScaler, which standardize features by removing the mean and scaling to unit variance; normalize, that normalizes vectors to unit norm and PCA_sklearn, it provides an implementation of the PCA algorithm.
- scipy: provides algorithms for optimization, integration, statistics and many other classes of problems. In this post we imported the function multivariate_normal, it enables the generation of random samples from a multivariate normal distribution.
- matplotlib: It's a comprehensive library for creating static, animated, and interactive visualizations.
Defining Functions
We define two functions: createDataset and PCA
# Create bivariate dataset
def createDataset(cov, mean, n_points, seed = 101):
# Defining dataset
distr = multivariate_normal(cov = cov, mean = mean, seed = seed)
X = distr.rvs(size = n_points)
# Scaling dataset
sc = StandardScaler()
X_scaled = sc.fit_transform(X)
return X_scaled
# PCA algorithm
def PCA(X, n_components):
# Calculating matrix of covariance
cov_matrix = np.cov(X.T)
# Eigenvectors and eigenvalues
eigenvalues, eigenvectors = eig(cov_matrix)
sort = np.argsort(eigenvalues)[::-1]
eigvalues_sort = eigenvalues[sort]
# Principal Components
eigvectors_sort = normalize(eigenvectors.T[sort], axis = 0)
# Matrix of transformation
transformation_matrix = eigvectors_sort.T
# Applying transformation to original dataset
reduced_dataset = X @ transformation_matrix[:,0:n_components]
return reduced_dataset, eigvectors_sort
The createDataset function generates a bivariate dataset with a multivariate normal distribution, taking the following parameters: covariance matrix (determines the shape and orientation of the distribution), mean vector (specifies the center of the distribution), number of points to generate and seed for the random number generator (optional). This function also standardizes the dataset using the StandardScaler function to ensure that have zero mean and unit variance.
The PCA function takes the input dataset X and the number of components n_components as parameters. It follows the steps early explained to perform the Principal Component Analysis algorithm and returns the reduced dataset and the sorted eigenvectors (principal components).
Generating Dataset
We generate a synthetic dataset X of 350 points using the createDataset function, and also defined one as the number of components for the PCA algorithm.
# Choosing parameters
cov = np.array([[1, 0.9], [0.9, 1]])
mean = np.array([0,0])
n_points = 350
n_components = 1
# Creating dataset
X = createDataset(cov, mean, n_points)
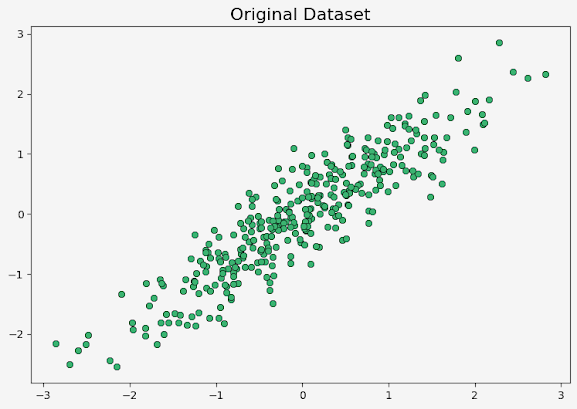
Plotting the original dataset
The matplotlib is used to create a scatter plot of the original dataset X.
# Plotting original dataset
plt.rcParams['font.size'] = '10'
f, axs = plt.subplots(1,1, figsize=(7,7), facecolor='#F5F5F5')
axs.set_facecolor('#F5F5F5')
axs.plot(X[:, 0], X[:, 1],'o', c='mediumseagreen',markeredgewidth = 0.5,markeredgecolor = 'black')
axs.set_title("Original Dataset", fontsize="16")
plt.show()
Applying PCA algorithm
Here we apply the PCA algorithm to the dataset X using the PCA function. It retains only 1 principal component (n_components). The reduced dataset and the corresponding eigenvectors are stored in the variables reduced_dataset and components, respectively.
# Applying PCA algorithm
reduced_dataset, components = PCA(X, n_components)
Another alternative is to use the PCA model from the library sklearn
# Applying PCA algorithm from sklearn
pca_sklearn = PCA_sklearn(n_components=1)
pca_sklearn.fit(X)
reduced_dataset = pca_sklearn.transform(X)
Plotting the PCA result (reduced dataset)
We create a scatter plot to visualize the reduced dataset after applying PCA algorithm. It plots the values from the reduced dataset on the x-axis. All y-values are 0 because the dataset has only one dimension, which corresponds to x-axis in this visualization.
# Plotting PCA result (reduced dataset with only one feature)
f, axs = plt.subplots(1,1, figsize=(9,5), facecolor='#F5F5F5')
axs.set_facecolor('#F5F5F5')
axs.scatter(reduced_dataset[:,0],len(reduced_dataset)*[0],s=200, zorder = -1, color="mediumseagreen", alpha = 0.3)
axs.set_title("Reduced dataset with only one feature", fontsize="16")
plt.show()
The result is:
Bonus: Visualizing the Principal Components
We create a scatter plot to visualize the directions of the principal components. The points of the original data set are plotted as green circles with transparency along with the two principal components as vectors (arrows), for this we use the components variable obtained from the PCA function.
# Plotting principal components
origin = np.array([[0, 0],[0, 0]]) # origin point
components_scaled = components * np.array([[1.0], [0.5]]) # scaled principal components
f, axs = plt.subplots(1,1, figsize=(7,7), facecolor='#F5F5F5')
axs.set_facecolor('#F5F5F5')
axs.scatter(X[:, 0], X[:, 1],s=50, zorder = -1, color="mediumseagreen", alpha = 0.3)
axs.quiver(*origin, components_scaled[:,0], components_scaled[:,1], color=['mediumblue','firebrick'], scale=3, headwidth = 4, width = 0.011)
axs.set_title("Principal Components Visualization", fontsize="16")
plt.show()
By following these steps, the code generates a bivariate dataset, applies PCA, and visualizes the original dataset, principal components, and the reduced dataset.








0 comments:
Post a Comment